
Welcome to NADCdb
Non-AIDS-defining cancers (NADC) have become a major cause of non-AIDS-related morbidity and mortality in people living with HIV (PLWH). However, the high-risk nature of HIV leads to difficulties in sample acquisition and data scarcity, posing significant challenges to pathological and clinical research on NADC.
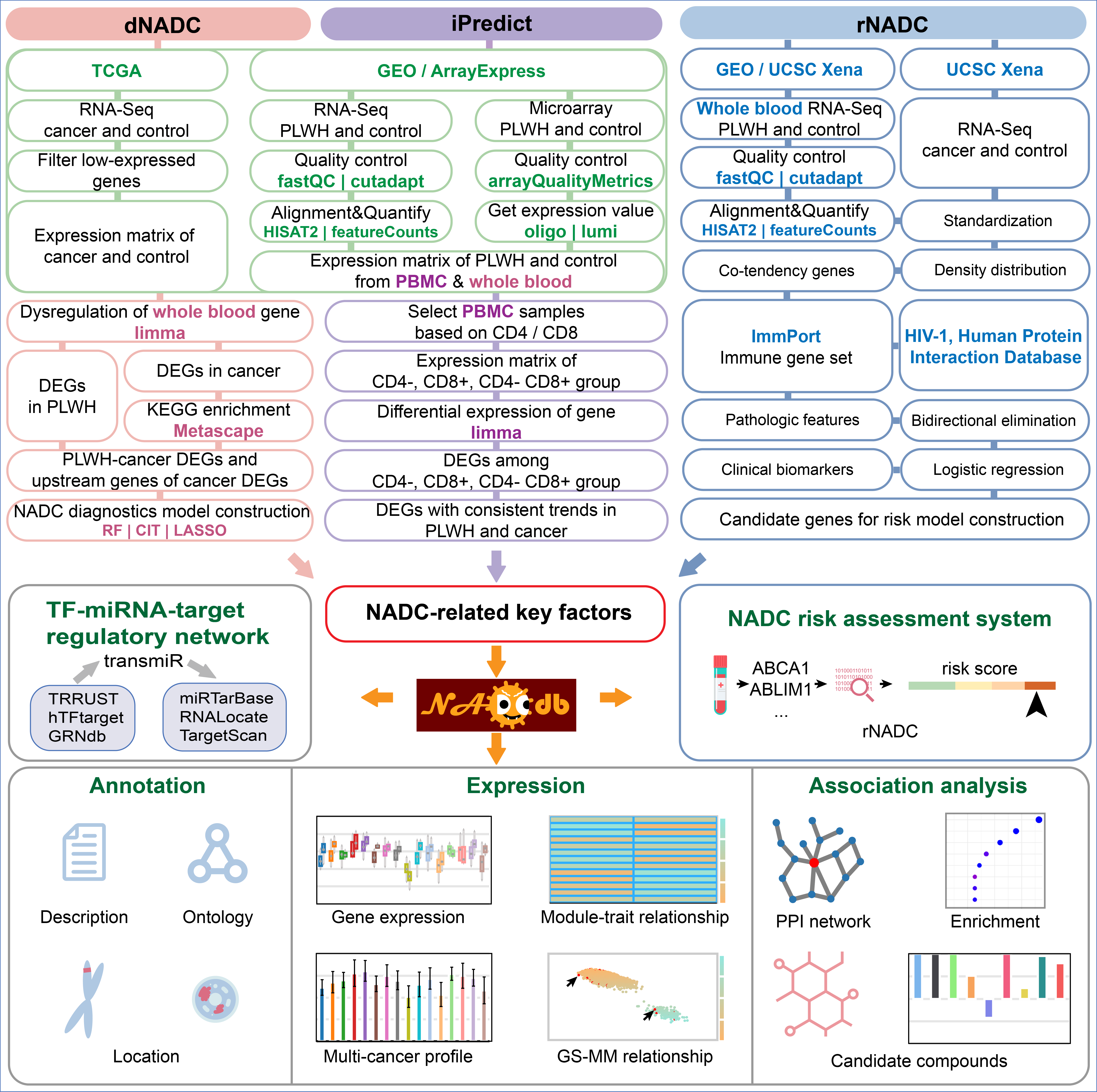
NADCdb is the first database dedicated to NADC pathological research. To overcome data scarcity, we innovatively employed a joint analysis strategy, deeply integrating and analyzing transcriptomic data from tens of thousands of PLWH and cancer patients, systematically identifying potential key regulators for 23 NADCs.
We developed three core models based on theories of immunosuppression, chronic inflammation, carcinogenic viral infection and oncogenic effects of HIV. rNADC was the first to utilize genes with abnormal expression trends as candidates, employing bidirectional stepwise regression and logistic regression to assess the risk of PLWH developing 21 NADCs. dNADC integrated patients' dysregulated genes and their regulatory networks, using Random Forest (RF) and Conditional Inference Trees (CIT) to identify key drivers influencing NADC progression with an accuracy exceeding 75%. iPredict identified 1,905 key immunobiomarkers for 16 NADCs based on distinct immune statuses of patients.
In summary, NADCdb serves as a novel, centralized resource that integrates data and provides analytical frameworks, offering fresh perspectives and a valuable platform for the scientific exploration of NADCs.
Workflow of NADCdb

NADCdb Features
NADCdb was released in Jnu, 2025.
- HIV samples: 205 ART samples, 147 non-ART samples, 241 healthy control samples.
- Cancer samples: 8,330 tumor tissue samples and 3563 paracancerous tissue of 23 cancers.
- Features: Immune indicators, inflammation indicators, HIV-human protein interaction and the common pathogenic pathway.
- Machine learning: Random forest (RF), Conditional inference tree (CIT), Least absolute shrinkage and selection operator (LASSO), Logistic regression
- Functions: GO, KEGG, Reactome, WikiPathways, Hallmark Gene Sets, BioCarta Gene Sets, Kinase Classe, Protein Functions, Subcellular Localization, Secretory Protein, PathogenicLoF, dbGap, GWAS, Human Phenotype Ontology, Drug, Disease and Gene Associations (DisGeNET), and Disease Ontology (DO), Tissue Specificity and Expression Patterns in Normal Tissues or Cancers.
- Associated analysis: PPI, WGCNA, CMap
- Associated factors: miRNA targets, transcription factors