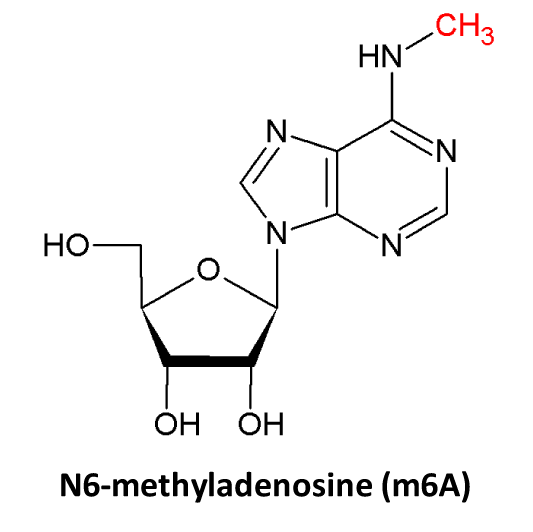
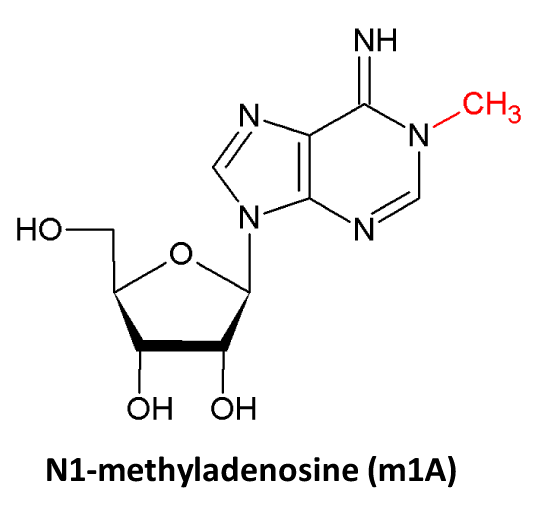
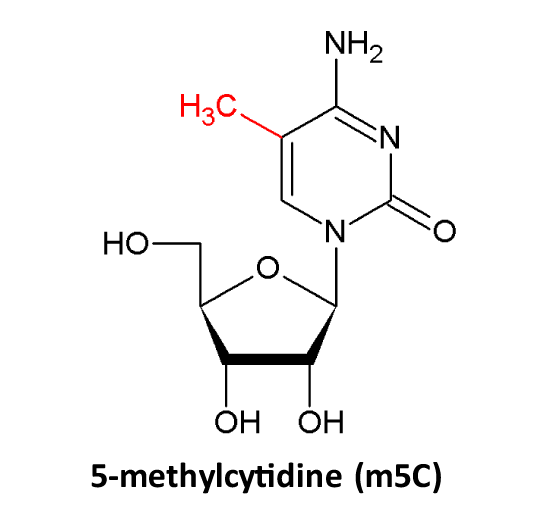
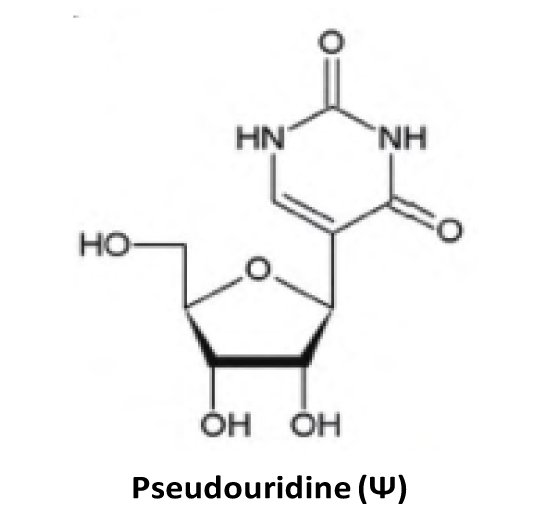
♥ Detailed notes are provided for each RNA modification site:
- Genome annotations include genome coordinates, gene biotypes, gene features, sequence;
- PWM Score is employed to assess the accuracy of m6A sites and ranges from 0 to 5, where a higher value indicates a more precise site. In RMBase, m6A modification sites are filtered using the criteria of support experimental number >= 2 and PWM score >= 3;
- Cell lines or tissues;
- Evolutionary conservation of mammalian m6A modifications is determined by liftOver or phyloP;
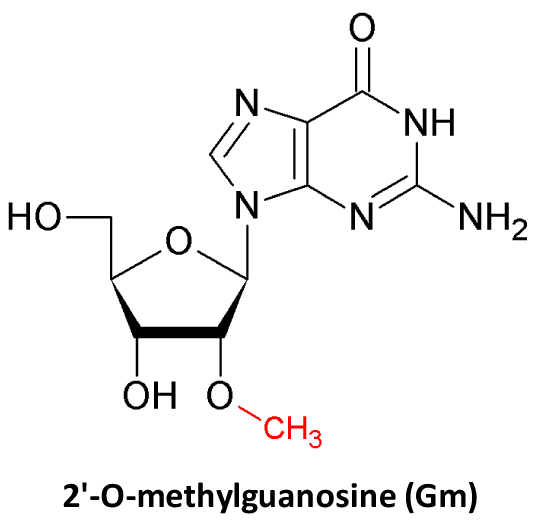
- snoRNA is used to annotate the snoRNA-guided 2'-O-Me (Nm) and Pseudo (Ψ), which can form a snoRNA-target pairing interactions;
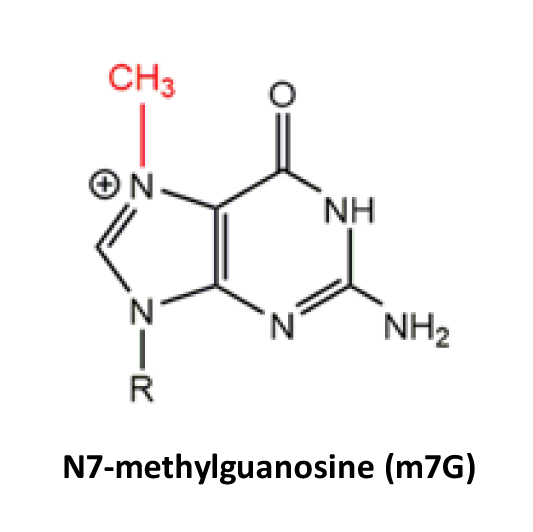
- Writer is applied to annotate RNA modification sites catalyzed by enzymes. e.g. METTL3 regulates the installation of m6A.
The Statistics of high accuracy RNA Modification sites
| Species | Assembly | m6A | m1A | m5C | pseudo | Nm | m7G | RNA editing | ac4C | Other | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | hg38 | 234464 | 1162 | 46025 | 15078 | 8490 | 919 | 144122 | 1861 | 1484 | 453605 |
| Mus musculus | mm10 | 220022 | 91 | 47839 | 22518 | 1193 | 55 | 8864 | 0 | 950 | 301532 |
| Macaca Mulatta | rheMac8 | 16699 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 16699 |
| Pan Troglodytes | panTro5 | 27828 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 27828 |
| Rattus | rn6 | 43880 | 75 | 118 | 1317 | 3418 | 46 | 10 | 24 | 977 | 49865 |
| Sus Scrofa | susScr11 | 62116 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 62116 |
| Danio Rerio | danRer11 | 5071 | 0 | 0 | 5 | 40 | 0 | 0 | 0 | 0 | 5116 |
| Arabidopsis thaliana | TAIR10 | 35373 | 0 | 0 | 76 | 286 | 0 | 0 | 0 | 0 | 35735 |
| Saccharomyces cerevisiae | sacCer3 | 24486 | 151 | 231 | 2147 | 263 | 75 | 66 | 120 | 1767 | 29306 |
| Drosophila melanogaster | BDGP6 | 56 | 78 | 46 | 614 | 559 | 47 | 5049 | 13 | 494 | 6956 |
| Pseudomonas aeruginosa | ASM676v1 | 1560 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1560 |
| Escherichia coli | ASM584v2 | 243 | 0 | 21 | 206 | 55 | 66 | 4 | 2 | 508 | 1105 |
| Schizosaccharomyces pombe | ASM294v2 | 1 | 8 | 8 | 41 | 18 | 9 | 0 | 0 | 87 | 172 |
| Bacillus subtilis | ASM69118v1 | 15 | 6 | 0 | 99 | 3 | 39 | 0 | 0 | 185 | 347 |
| Bombyx mori | ASM15162v1 | 0 | 82 | 178 | 185 | 95 | 33 | 0 | 0 | 453 | 1026 |
| Bos taurus | bosTau9 | 0 | 127 | 94 | 381 | 105 | 29 | 0 | 16 | 770 | 1522 |
| Brassica napus | AST_PRJEB5043v1 | 0 | 49 | 0 | 175 | 128 | 59 | 0 | 0 | 425 | 836 |
| Caenorhabditis elegans | WBcel235 (ce11) | 0 | 0 | 0 | 296 | 151 | 0 | 18 | 0 | 106 | 571 |
| Candida albicans | Cand_albi12Cv2 | 0 | 1 | 1 | 3 | 1 | 0 | 0 | 1 | 6 | 13 |
| Chlamydomonas reinhardtii | CHLRE3 | 0 | 0 | 2 | 30 | 34 | 0 | 0 | 0 | 9 | 75 |
| Clostridium acetobutylicum | ASM78585v1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 2 |
| Cucumis sativus | ASM407v2 | 0 | 12 | 14 | 34 | 0 | 0 | 0 | 0 | 105 | 165 |
| Enterobacteria phage T4 | ViralProj14044 | 0 | 0 | 0 | 14 | 6 | 3 | 0 | 0 | 39 | 62 |
| Enterobacteria phage T5 | ASM292140v1 | 1 | 0 | 0 | 9 | 1 | 2 | 0 | 0 | 10 | 23 |
| Fusarium graminearum | ASM24013v3 | 0 | 0 | 0 | 0 | 0 | 0 | 48493 | 0 | 0 | 48493 |
| Fusarium verticillioides | ASM14955v1 | 0 | 0 | 0 | 0 | 0 | 0 | 5227 | 0 | 0 | 5227 |
| Gallus gallus | GRCg6a | 0 | 4 | 0 | 36 | 55 | 2 | 0 | 0 | 37 | 134 |
| Geobacillus stearothermophilus | ASM74998v1 | 1 | 3 | 0 | 11 | 5 | 3 | 0 | 0 | 25 | 48 |
| Glycine max | glyMax2 | 0 | 0 | 0 | 4 | 1 | 1 | 0 | 0 | 7 | 13 |
| Halobacterium salinarum | ASM680v1 | 0 | 0 | 10 | 17 | 10 | 0 | 0 | 0 | 31 | 68 |
| Halococcus morrhuae | ASM33669v1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 |
| Haloferax volcanii | ASM2568v1 | 0 | 0 | 51 | 79 | 51 | 0 | 0 | 5 | 167 | 353 |
| Lactococcus lactis | ASM76111v1 | 11 | 2 | 0 | 0 | 0 | 30 | 0 | 1 | 64 | 108 |
| Mesocricetus auratus | MesAur1 | 0 | 2 | 0 | 5 | 0 | 0 | 0 | 0 | 9 | 16 |
| Mycobacterium smegmatis | ASM76770v1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 4 |
| Mycoplasma capricolum | ASM83033v1 | 12 | 10 | 0 | 38 | 2 | 17 | 0 | 0 | 78 | 157 |
| Mycoplasma mycoides | ASM25307v1 | 4 | 1 | 0 | 4 | 0 | 3 | 0 | 0 | 14 | 26 |
| Neurospora crassa | NC12 | 0 | 22 | 14 | 36 | 0 | 10 | 47345 | 0 | 134 | 47561 |
| Nicotiana tabacum | Ntab_TN90 | 0 | 25 | 0 | 134 | 123 | 0 | 0 | 0 | 296 | 578 |
| Oryctolagus cuniculus | OryCun2 | 0 | 88 | 98 | 225 | 46 | 70 | 0 | 0 | 493 | 1020 |
| Ovis aries | Oarv3 | 0 | 24 | 35 | 35 | 0 | 15 | 0 | 0 | 132 | 241 |
| Phaseolus vulgaris | PhaVulg1 | 0 | 35 | 19 | 78 | 42 | 15 | 0 | 12 | 256 | 457 |
| Pichia jadinii | Cybja1 | 0 | 40 | 43 | 108 | 31 | 17 | 0 | 7 | 311 | 557 |
| Pyrococcus abyssi | ASM19593v2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Rhodospirillum rubrum | ASM1308v1 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 4 | 7 |
| Salmonella typhimurium | ASM553737v1 | 0 | 0 | 0 | 19 | 7 | 3 | 0 | 0 | 42 | 71 |
| Solanum tuberosum | SolTub3 | 0 | 18 | 14 | 46 | 17 | 0 | 0 | 16 | 117 | 228 |
| Spinacia oleracea | ASM200726v1 | 0 | 11 | 0 | 89 | 81 | 17 | 0 | 0 | 251 | 449 |
| Spiroplasma citri | ASM188685v1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 3 | 7 |
| Streptomyces coelicolor A3(2) | ASM20383v1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 2 |
| Streptomyces griseus | ASM93222v1 | 0 | 26 | 0 | 4 | 25 | 18 | 0 | 0 | 48 | 121 |
| Sulfolobus acidocaldarius | ASM1228v1 | 0 | 1 | 1 | 0 | 4 | 0 | 0 | 0 | 1 | 7 |
| Sulfolobus solfataricus | ASM96839v1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 24 | 24 |
| Synechococcus elongatus PCC 6301 | ASM1006v1 | 0 | 0 | 0 | 8 | 1 | 1 | 0 | 0 | 11 | 21 |
| Synechococcus sp. PCC 7002 | ASM1948v1 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 0 | 5 | 9 |
| Synechocystis sp. | ASM34078v1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 3 | 4 |
| Tetrahymena thermophila | JCVI_TTA1_2 | 0 | 50 | 49 | 165 | 81 | 0 | 0 | 0 | 205 | 550 |
| Thermoplasma acidophilum | ASM19591v1 | 0 | 4 | 0 | 10 | 4 | 2 | 0 | 0 | 15 | 35 |
| Thermotoga maritima | ASM97857v1 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 5 | 8 |
| Thermus thermophilus | ASM812v1 | 0 | 4 | 12 | 19 | 17 | 6 | 0 | 0 | 28 | 86 |
| Xenopus laevis | Xenopus_laevisv2 | 0 | 55 | 86 | 104 | 46 | 35 | 0 | 0 | 270 | 596 |
| Zea mays | RefGenv4 | 0 | 0 | 0 | 15 | 9 | 0 | 0 | 0 | 33 | 57 |