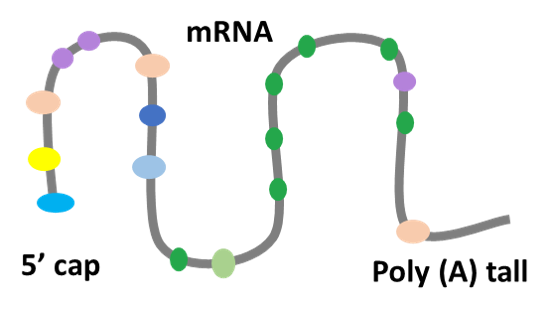
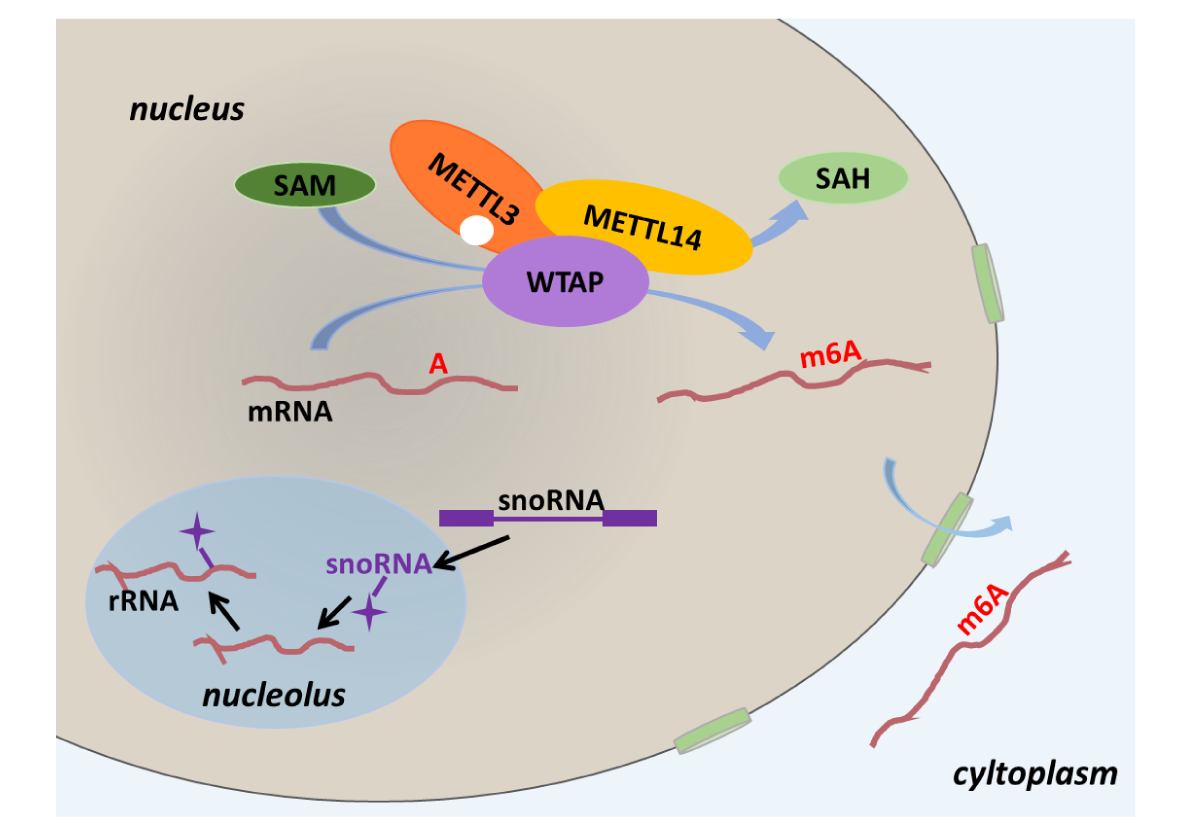
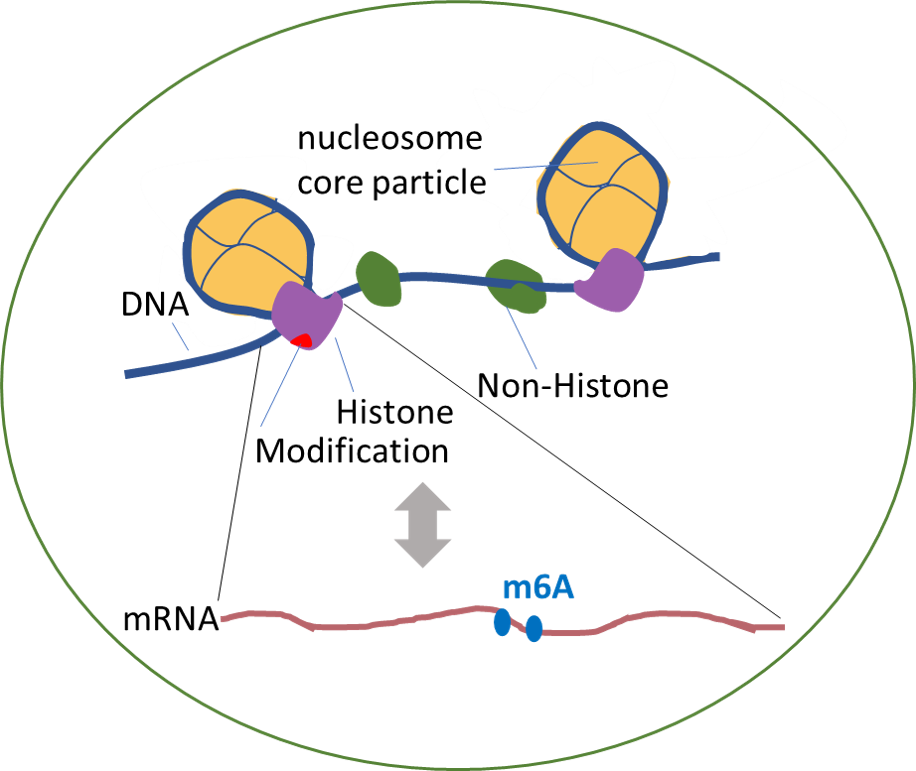
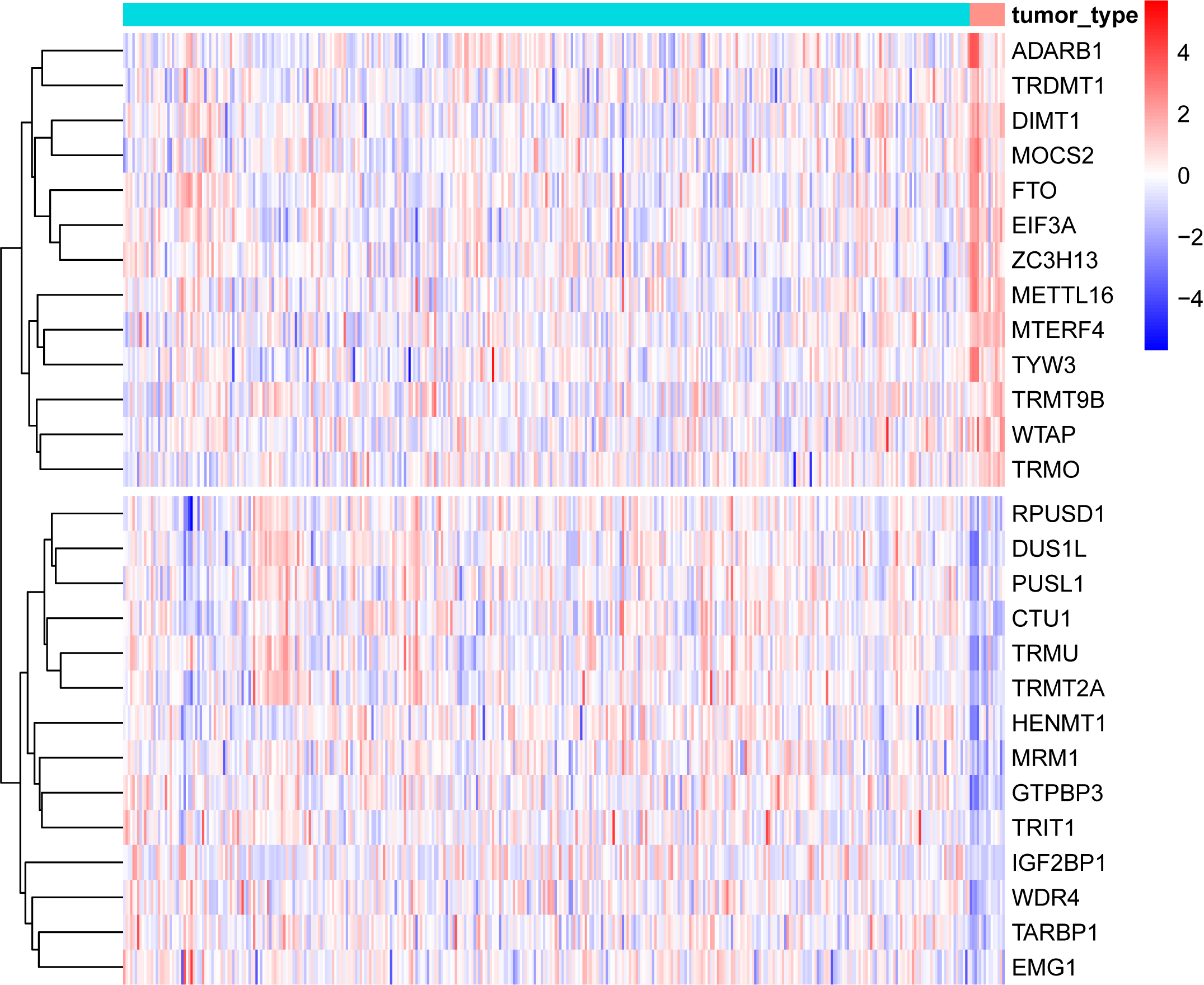
Welcome to RMBase v3.0!
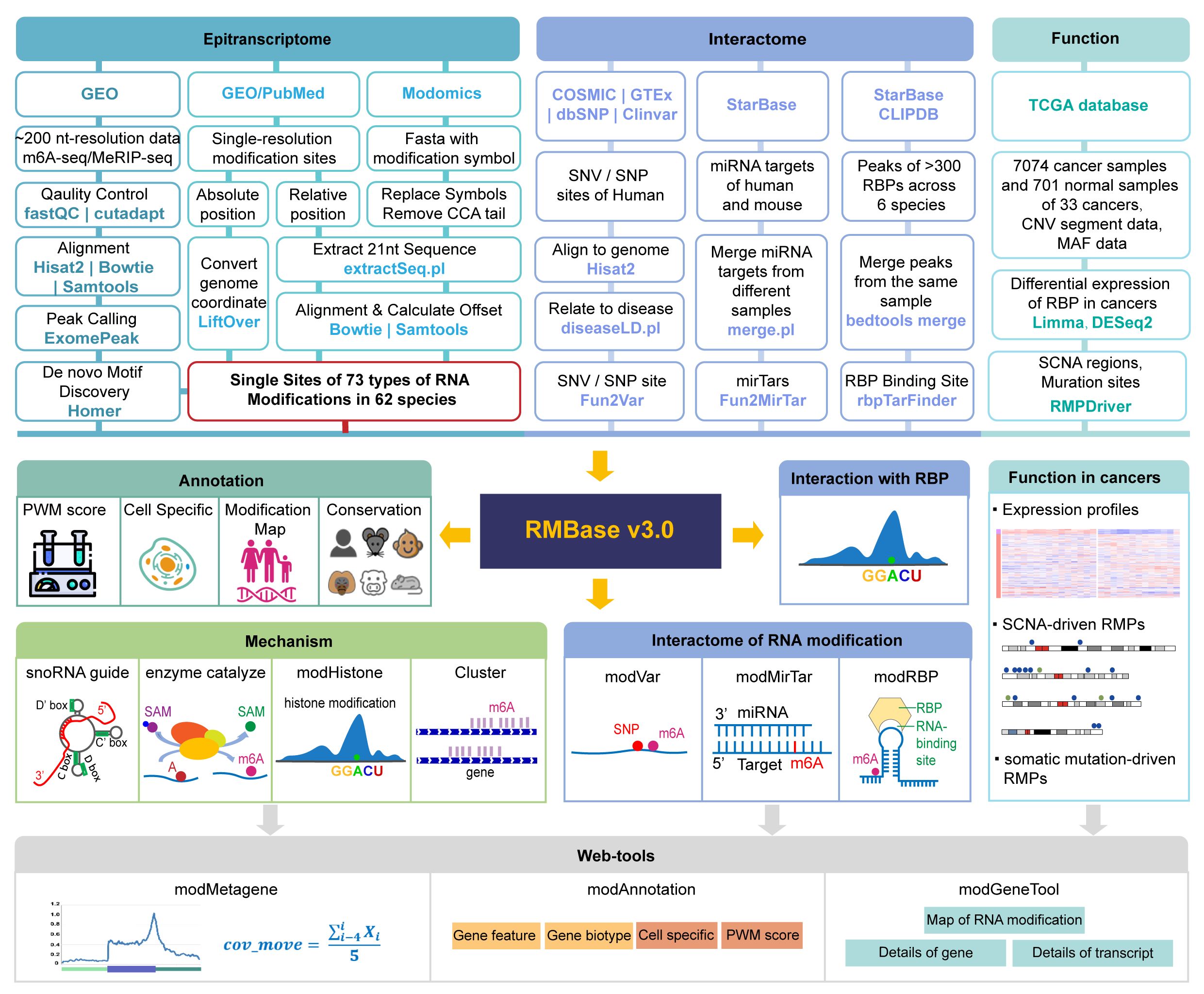
Although over 170 chemical modifications have been identified, their prevalence, mechanism and function remain largely unknown. To enable integrated analysis of diverse RNA modification profiles, we have developed RMBase v3.0, also call ENCORE (The Encyclopedia of RNA Epitranscriptome), a comprehensive platform consisting of eight modules. These modules facilitate the exploration of transcriptome-wide landscape, biogenesis, interactome and functions of RNA modifications. By mining thousands of epitranscriptome datasets with novel pipelines, the ‘RNA Modifications’ module reveals the map of 73 RNA modifications of 62 species. the ‘Genes’ module allows to retrieve RNA modification profiles and clusters by gene and transcript. The ‘Mechanisms’ module explores 23 382 enzyme-catalyzed or snoRNA-guided modified sites to elucidate their biogenesis mechanisms. The ‘Co-localization’ module systematically formulates potential correlations between 14 histone modifications and 6 RNA modifications in various cell-lines. The ‘RMP’ module investigates the differential expression profiles of 146 RNA-modifying proteins (RMPs) in 18 types of cancers. The ‘Interactome’ integrates the interactional relationships between 73 RNA modifications with RBP binding events, miRNA targets and SNPs. The ‘Motif’ illuminates the enriched motifs for 11 types of RNA modifications identified from epitranscriptome datasets. The ‘Tools’ introduces a novel web-based ‘modGeneTool’ for annotating modifications. Overall, RMBase v3.0 provides various resources and tools for studying RNA modifications.
The database idetified 1,074,100 RNA modification sites among 73 type of RNA modifications in 62 .
RMBase v3.0 Release Note (Oct 2023):
- Species: 9 clades, 62 species.
- Samples: 828 wild type datasets.
- RNA Modification: 73 types.
- Tools for RNA modifications: 3 tools.
- Explore the pseudo sites guided by known and orphan snoRNAs. These pseudo sites occur on mRNA, rRNA, snRNA, tRNA, lncRNA and repeatMasker.
- Detect the RNA modification clusters in genome wide.
- Decipher the associated functions of RNA modifications with RBP, miRNA target, SNP, and SNV.
- Identify novel m6A readers.
- Explore the potential associations (co-localization) of histone modifications on RNA modifications.
- Investigate the biological functions of RNA modifications in diverse cancers.
How to cite:
- Xuan JJ, et al. RMBase v3.0: decode the landscape, mechanisms and functions of RNA modifications. Nucleic Acids Res. 2023 Nov 13.
- Xuan JJ, et al. RMBase v2.0: Deciphering the Map of RNA Modifications from Epitranscriptome Sequencing Data. Nucleic Acids Res. 2018 Jan 4;46(D1):D327-34.
- Sun WJ, et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016 Jan 4;44(D1):D259-65.